News
Newswise —
Researchers from the Charles E. Schmidt College of Medicine at Florida Atlantic University have engineered endogenous protein inhibitors of protein-degrading enzymes as an alternative approach to synthetic inhibitors for potentially treating cancer and other diseases. Results of their study, titled “Thermodynamics of Selectivity in N-TIMP/MMP Interactions,” were recently published in the Journal of Biological Chemistry.They have been investigating a group of four proteins called tissue inhibitors of metalloproteinases (TIMPs) that inhibit and control the activities of a group of 23 enzymes called matrix metalloproteinases (MMPs). Pharmaceutical companies have previously investigated the possible use of synthetic MMP inhibitors for treating cancer and other diseases, but they failed in clinical trials due to side effects, most likely because they were insufficiently specific and inhibited MMPs needed for normal physiological processes.
“There are 23 MMPs in humans that degrade or hydrolyze all of the protein components of extracellular matrices such as cartilage, bone, and basement membranes,” said Keith Brew, Ph.D., a professor of biomedical science in FAU’s College of Medicine. “If the regulation of their activities fails, MMPs can damage cells and extracellular structures, and their unregulated activities that are linked to serious diseases including osteoarthritis, heart disease, and cancer metastasis.”
The TIMPs developed by Brew were designed to make them specific inhibitors of selected MMP targets to potentially treat osteoarthritis and cancer.
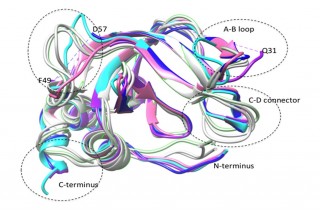
“In our study, we have been able to determine the thermodynamic profiles for the interactions of normal and mutant TIMPs with three MMPs, so that we can develop an understanding of the relationship between structural changes produced by the mutations and the changes in their abilities to inhibit different MMPs,” said Brew.
To accomplish this, the researchers used a technique called “isothermal titration calorimetry” (ITC), which measures the minute amounts of heat that are given out or absorbed when two molecules interact together (the enthalpy of binding). When they measured how this varies as they increased the quantities of one protein under different conditions of temperature and solvent, they were able to determine the enthalpy change (heat absorbed or released), entropy change (a measure of dynamics in the proteins and their surroundings) and heat capacity change, enabling them to separately estimate the changes in protein and solvent dynamics.
Unlike most other protein-to-protein interactions, they found that TIMP/MMP interactions are driven by entropy increases, and not by enthalpy (which derives from specific interactions between chemical groups in the interacting molecules). They found that proportions of entropy coming from internal dynamics in the protein and from the solvent varies for different interactions, and is affected by TIMP mutations, but that the MMP component has a major role in determining the proportions.
“When we think of two proteins binding together, we have to consider that they are dynamic molecules, each consisting of a population of interconverting three-dimensional structures,” said Brew. “Complex formation involves the selection of subpopulations that are compatible with each other in structures and dynamics, a process called ‘conformer selection.’ And, it appears that the MMP component has a major role in this process making the rational design of selective mutants difficult.”
Co-authors of the study also include FAU’s College of Medicine and Charles E. Schmidt College of Science Ph.D. students Haiyin Zou and Ying Wu.
This research is supported by the National Institutes of Health (NIH), grant AR40994.- FAU -
About Florida Atlantic University:Florida Atlantic University, established in 1961, officially opened its doors in 1964 as the fifth public university in Florida. Today, the University, with an annual economic impact of $6.3 billion, serves more than 30,000 undergraduate and graduate students at sites throughout its six-county service region in southeast Florida. FAU’s world-class teaching and research faculty serves students through 10 colleges: the Dorothy F. Schmidt College of Arts and Letters, the College of Business, the College for Design and Social Inquiry, the College of Education, the College of Engineering and Computer Science, the Graduate College, the Harriet L. Wilkes Honors College, the Charles E. Schmidt College of Medicine, the Christine E. Lynn College of Nursing and the Charles E. Schmidt College of Science. FAU is ranked as a High Research Activity institution by the Carnegie Foundation for the Advancement of Teaching. The University is placing special focus on the rapid development of critical areas that form the basis of its strategic plan: Healthy aging, biotech, coastal and marine issues, neuroscience, regenerative medicine, informatics, lifespan and the environment. These areas provide opportunities for faculty and students to build upon FAU’s existing strengths in research and scholarship. For more information, visit www.fau.edu.
Newswise —
Researchers from University College London have discovered that the widely used antiinflammatory drug HUMIRA doesn’t just work by inhibiting its target protein, TNF, but by enhancing a particular function of TNF in rheumatoid arthritis patients. The study, “Anti-TNF drives regulatory T cell expansion by paradoxically promoting membrane TNF–TNF-RII binding in rheumatoid arthritis,” which will be published online June 6 in The Journal of Experimental Medicine, may help explain the divergent efficacies of different TNF-targeting drugs.
TNF is an important inflammatory molecule produced by the immune system, and TNF inhibitors are commonly used to treat inflammatory diseases such as rheumatoid arthritis and Crohn’s disease. One such inhibitor is the monoclonal anti-TNF antibody adalimumab, commonly marketed under the brand name HUMIRA. Michael Ehrenstein and colleagues at University College London previously found that treating rheumatoid arthritis patients with adalimumab increased the number of regulatory T cells capable of suppressing inflammation. Researchers presumed that this was because, in the absence of adalimumab, TNF blocks the development of these regulatory T cells. Mysteriously, however, another TNF inhibitor, etanercept, doesn’t induce the formation of these inflammation-suppressing T cells in rheumatoid arthritis patients.
Ehrenstein and his colleague, Dao Xuan Nguyen, now report that, unlike etanercept, adalimumab actually enhances the ability of TNF to induce the formation of antiinflammatory T cells. In particular, Nguyen and Ehrenstein discovered that adalimumab increases the expression of TNF on the surface of patient monocytes and promotes the association of these TNF molecules with receptor proteins on the surface of regulatory T cells. This activates a signaling pathway that endows the T cells with the capacity to suppress inflammation.
Adalimumab may therefore block the proinflammatory functions of soluble TNF secreted by immune cells, while augmenting the activity of membrane-bound TNF that ultimately helps to resolve inflammation. “These results highlight how a treatment that targets a pivotal inflammatory cytokine not only preserves but actually boosts the pro-resolution forces driven by that pathway, thereby introducing a novel therapeutic paradigm,” Ehrenstein says.
Nguyen, D.X., and M.R. Ehrenstein. 2016. J Exp. Med. http://dx.doi.org/10.1084/jem.20151255
# # #
Newswise — MADISON, Wis. —
A new mouse model, developed by researchers at the University of Wisconsin–Madison, is the first to show that when more of a specific biological molecule moves between different parts of nerve cells in the mouse brain, it can lead to behaviors that resembles some aspects of autism spectrum disorder (ASD) in humans.
This biological molecule, called acetyl-CoA, is a major part of the process cells use to make energy from food. It’s also used within cells to tag different proteins, which influences where and how they function. Local concentrations of acetyl-CoA and its movement, or flux, between different areas within cells is tightly regulated.
“We show, for the very first time, that changes in acetyl-CoA flux, and not just changes in its levels, in individual neurons can affect neuronal activity,” says Luigi Puglielli, a professor in the Department of Medicine of the UW-Madison School of Medicine and Public Health and the UW’s Waisman Center.
In the study, published this week in The Journal of Experimental Medicine, the researchers engineered mice to make the human version of a protein that ferries acetyl-CoA into a specific compartment within cells. Mouse models of ASD can help scientists understand the molecular basis of the disorder.
Previous studies revealed that mutations in this ferrying protein, which is called AT-1, are associated with spastic paraplegia, severe developmental delays and autism spectrum disorder in humans. But how mutations in AT-1 are linked to these developmental disorders was unknown.
The current study showed that changes in the amount of AT-1 in nerve cells can profoundly influence how much acetyl-CoA is found in different areas within those cells. When AT-1 levels are high, as is the case in the brains of the mice with the human AT-1 protein, increased movement of acetyl-CoA into specific areas within cells sets off a chain reaction of consequences that the researchers think ultimately leads to the mice showing autism-like behaviors.
“We could call AT-1 a ‘master regulator’ of intracellular acetyl-CoA flux, which, in turn, can be said to be a master regulator of essential neuronal functions,” says Puglielli.
In the brains of mice with human AT-1, atypical localization of acetyl-CoA in the nerve cells causes a slew of more than 400 genes to become dysregulated and pump out higher levels of proteins. Several of these proteins play important roles in regulating both the growth of neurons and how nerve impulses travel through them.
The global changes in protein levels caused by manipulating these master regulators leads to significant changes in what nerve cells look like and how they function in these mice. For instance, the ends of the nerve cells become more branched and spiny and their ability to mediate typical learning and memory formation is compromised.
Puglielli and his colleagues think these changes in how the nerve cells look and function ultimately caused the AT-1 mice to behave atypically, in ways that resemble aspects of ASD in humans.
“We need to be able to modify genetic, molecular and biochemical aspects of the disorder,” says Puglielli. “These sort of manipulations and studies cannot be performed in humans, hence the need to develop and study mouse models.”
While mouse models can provide vital information about human disorders, such as ASD, the researchers urge caution while interpreting findings.
“ASD is difficult to define in humans and there are different behaviors that we globally include under the umbrella of autism,” says Puglielli. “If it is difficult to define autism — a human disorder — in humans, you can imagine how much more difficult it is to define in mice.”
Puglielli and his colleagues are now looking at other proteins that regulate acetyl-CoA movement within cells. “Mutations in these proteins are also associated with different disorders, including ASD and intellectual disability,” he says. “A comprehensive analysis of the functions of these proteins will help us dissect more aspects of how acetyl-CoA flux is relevant to ASD.”
—Adityarup “Rup” Chakravorty
Newswise —
Drug-cue associations can have a powerful influence over individuals with drug and alcohol use disorders, often leading to relapse in those attempting to stay abstinent. Few studies have investigated how drugs affect learning or memory for drug-associated stimuli in humans. This study examined the direct effects of alcohol on memory for images of alcohol-related beverages, such as beer bottles or liquor glasses, or neutral beverages, such as water bottles or soda cans, in social drinkers.
Researchers assigned subjects to one of three conditions: one group (n=20) received an intoxicating dose of alcohol (0.8 g/kg) before viewing visual images, called the Encoding condition; a second group (n=20) received the same amount of alcohol immediately after viewing them, called the Consolidation condition. A third group (n=19) received a placebo both before and after viewing the images, called the Control condition. Memory retrieval was tested exactly 48 hours later, in a drug-free state.
Results indicate that alcohol impairs memory in the Encoding condition and enhances memory in the Consolidation condition. However, individual differences in sensitivity to alcohol’s positive rewarding effects are associated with a greater tendency to remember alcohol-related environmental stimuli encountered while intoxicated. In other words, these individuals may form stronger memory associations with alcohol-related stimuli, which may then strengthen their drinking behavior and/or place them at a greater risk of relapse if they decide to stop drinking.
Newswise —
Physicians have long used magnetic resonance imaging (MRI) to detect cancer but results of a University of California San Diego School of Medicine study describe the potential use of restriction spectrum imaging (RSI) as an imaging biomarker that enhances the ability of MRI to differentiate aggressive prostate cancer from low-grade or benign tumors and guide treatment and biopsy.
“Noninvasive imaging is used to detect disease, but RSI-MRI takes it a step further,” said David S. Karow, MD, PhD, assistant professor of radiology at UC San Diego School of Medicine and the study’s senior author. “We can predict the grade of a tumor sometimes without a biopsy of the prostate tissue. This is taking all that’s good about multi-parametric MRI and making it better.”
The addition of RSI to a pelvic MRI added between 2.5 to 5 minutes to scanning time making it a fast and highly accurate tool with decreased risk compared to contrast MRI which involves injecting patients with dye, said Karow.
In the study, published online June 1 in Clinical Cancer Research, the authors said RSI-MRI corrects for magnetic field distortions found in other imaging techniques and focuses upon water diffusion within tumor cells that exhibit a high nuclear volume fraction. By doing this, the ability of imaging to accurately plot a tumor’s location is increased and allows for differentiation between tumor grades. The higher the grade, the more aggressive the cancer. Patients can have more than one tumor with different grades, however. Karow said RSI-MRI can be used to guide treatment or biopsy to target the region of highest-grade cancer.
An early diagnosis of prostate cancer typically improves a patient’s prognosis. According to the National Cancer Institute, prostate cancer is the second leading cause of cancer death in men in the United States, with more than 26,000 estimated deaths this year and 180,890 new diagnoses predicted. The average age at the time of diagnosis is 66.
At UC San Diego Health, more than 1,000 patients have been imaged with RSI-MRI since 2014 and a subset have subsequently undergone MR-fused ultrasound guided prostate biopsy, said J. Kellogg Parsons, MD, MHS, UC San Diego School of Medicine associate professor of surgery and study co-author.
“Previously, we relied completely on systematic — but random — biopsies of the prostate to diagnose cancer, which has been the standard practice in our field for years. Now, we use RSI-MRI to precisely target specific areas of concern and enhance the accuracy of our diagnosis,” said Parsons, surgical oncologist at Moores Cancer Center at UC San Diego Health.
“Greater accuracy means improved care tailored to each individual patient. With RSI-MRI, we are better able to identify which cancers are more aggressive and require immediate treatment, and which ones are slow growing and can be safely observed as part of a program called active surveillance.”
Although this study focused on 10 patients, more than 2,700 discrete data points were evaluated. Next steps include introducing the technology to other hospitals and to study whether it can be used in isolation from other screening tools. In prior papers published in the journalsAbdominal Radiology and Prostate Cancer Prostatic Diseases, the same authors reported that RSI-MRI increases detection capability and can perform better than traditional multi-parametric MRI when used in isolation.
These data suggest that RSI-MRI could eventually serve as a stand-alone, non-contrast screening tool that would take 15 minutes compared to a normal contrast-enhanced exam lasting 40 to 60 minutes.
“What our evidence shows so far is the imaging benefit is coming from RSI-MRI,” said Karow. “I think this technique could become standard of care and mainstream for the vast majority of men who are at risk for prostate cancer. Full contrast MRI is expensive and risky for most men. This is the kind of exam that could be done on a routine clinical basis.”
Anders Dale, PhD, professor of radiology and neurosciences and co-director of the Multimodal Imaging Laboratory at UC San Diego, and Nate White, PhD, assistant professor of radiology, initially co-invented RSI-MRI to characterize aggressive brain tumors.
“RSI-MRI could be a transformational imaging technology for oncologists in the same way CT scans altered the way effects of treatment are quantitated from plain X-rays,” said Jonathan W. Simons, MD, Prostate Cancer Foundation president and Chief Executive Officer. “Based on the investigations at UC San Diego, this is a particular promise that needs more validation. Now testable is the hypothesis that RSI-MRI could identify oligometastatic prostate cancer that became curable through its identification by RSI-MRI.”
Additional study co-authors include: Ghiam Yamin, Natalie M. Schenker-Ahmed, Ahmed Shabaik, Dennis Adams, Hauke Bartsch, Joshua Kuperman, Nate White, Rebecca A. Rakow-Penner, Kevin McCammack, Christopher J. Kane, and Anders M. Dale, all at UC San Diego.
This research was funded, in part, by Department of Defense Prostate Cancer Research Program (W81XWH-13-1-0391), American Cancer Society Institutional Research grant (70-002), UCSD Clinician Scientist Program (5T32EB005970-07), UCSD School of Medicine Microscopy Core and NINDS P30 core grant (NS047101), General Electric Investigator Initiated Research Award (BOK92325), and the National Science Foundation (1430082).
###
Newswise —
Statistics show that some 15 million Americans don’t work the typical nine-to-five. These employees (or shift workers), who punch in for graveyard or rotating shifts, are more prone to numerous health hazards, from heart attacks to obesity, and now, new research, published in Endocrinology, shows shift work may also have serious implications for the brain.
“The body is synchronized to night and day by circadian rhythms—24-hour cycles controlled by internal biological clocks that tell our bodies when to sleep, when to eat and when to perform numerous physiological processes,” said David Earnest, Ph.D., professor in the Department of Neuroscience and Experimental Therapeutics at the Texas A&M Health Science Center College of Medicine. “A person on a shift work schedule, especially on rotating shifts, challenges, or confuses, their internal body clocks by having irregular sleep-wake patterns or meal times.”
According to Earnest, it’s not the longer hours—or the weird hours—necessarily that is the problem. Instead, it is the change in the timing of waking, sleeping and eating every few days that “unwinds” our body clocks and makes it difficult for them to maintain their natural, 24-hour cycle. When body clocks are disrupted, as they are when people go to bed and get up at radically different times every few days, there can be a major impact on health. Earnest and his colleagues have found that shift work can lead to more severe ischemic strokes, the leading cause of disability in the United States, which occur when blood flow is cut off to part of the brain.
Using an animal model, Earnest and his team, including colleague Farida Sohrabji, Ph.D., also a professor in the Department of Neuroscience and Experimental Therapeutics and director of the Women’s Health in Neuroscience Program, found that subjects on shift work schedules had more severe stroke outcomes, in terms of both brain damage and loss of sensation and limb movement than controls on regular 24-hour cycles of day and night.
Of interest, their study—supported by the American Heart Association—found that males and females show major differences in the degree to which the stroke was exacerbated by circadian rhythm disruption; in males, the gravity of stroke outcomes in response to shift work schedules was much worse than in females.
“These sex differences might be related to reproductive hormones. Young women are less likely to suffer strokes, as compared with men of a similar age, and when they do, the stroke outcomes are likely to be less severe. In females, estrogen is thought to be responsible for this greater degree of neuroprotection,” Sohrabji said. “Essentially, estrogen helps shield the brain in response to stroke.” However, older women approaching menopause show increasing incidence of ischemic stroke and poor prognosis for recovery, compared with men at the same age.
Some of Earnest’s previous work has shown that a high-fat diet can also alter the timing of internal body clocks, as well as dramatically increase inflammatory responses that can be a problem in cardio- and cerebrovascular disease (conditions caused by problems that affect the blood supply to the brain—which includes stroke).
“Next we would like to explore whether inflammation is a key link between circadian rhythm disruption and increased stroke severity,” Earnest said. “With this information, we may be able to identify therapeutic interventions that limit damage after a stroke in patients with a history of shift work.”
“This research has clear implications for shift workers with odd schedules, but probably extends to many of us who keep schedules that differ greatly from day-to-day, especially from weekdays to weekends,” Earnest added. “These irregular schedules can produce what is known as ‘social jet lag,’ which similarly unwinds our body clocks so they no longer keep accurate time, and thus can lead to the same effects on human health as shift work.”
An immediate impact of these studies on human health is that individuals in shift work-type professions should be monitored more closely and more frequently for cardio- and cerebrovascular disease and risk factors such as hypertension and obesity.
In the meantime, Earnest suggests that those with irregular sleeping patterns should at least try to maintain regular mealtimes, in addition to avoiding the usual cardiovascular risk factors like a high-fat diet, inactivity and tobacco use.
Newswise —
How does stress – which, among other things, causes our bodies to divert resources from non-essential functions – affect the basic exchange of materials that underlies our everyday life? Weizmann Institute of Science researchers investigated this question by looking at a receptor in the brains of mice, and they came up with a surprising answer. The findings, which recently appeared in Cell Metabolism, may in the future aid in developing better drugs for stress-related problems and eating disorders.
Dr. Yael Kuperman began this study as part of her doctoral research in the lab of Prof. Alon Chen of the Department of Neurobiology. Dr. Kuperman, presently a staff scientist in the Veterinary Resources Department, Prof. Chen, and research student Meira Weiss focused on an area of the brain called the hypothalamus, which has a number of functions, among them helping the body adjust to stressful situations, controlling hunger and satiety, and regulating blood glucose and energy production.
When stress hits, cells in the hypothalamus step up production of a receptor called CRFR1. It was known that this receptor contributes to the rapid activation of a stress-response sympathetic nerve network – increasing heart rate, for example. But since this area of the brain also regulates the body’s exchange of materials, the team thought that the CRFR1 receptor might play a role in this, as well.
Prof. Chen and his group characterized the cells in a certain area of the hypothalamus, finding that the receptor is expressed in around half of the cells that arouse appetite and suppress energy consumption. These cells comprise one of two main populations in the hypothalamus – the second promotes satiety and the burning of energy. “This was a bit of a surprise,” says Dr. Kuperman, “as we would instinctively expect the receptor to be expressed on the cells that suppress hunger.”
To continue investigating, the researchers removed the CRFR1 receptor in mice from just the cells that arouse appetite in the hypothalamus, and then observed how this affected the animals’ bodily functions. At first, the team did not see any significant changes, confirming that this receptor is saved for stressful situations. But when they exposed the mice to stress – cold or hunger – they got another surprise.
When exposed to cold, the sympathetic nervous system activates a unique type of fat called brown fat, which produces heat to maintain the body’s internal temperature. When the receptor was removed, the body temperature dropped dramatically – but only in the female mice. Their temperatures failed to stabilize even afterward the stressor was removed, while male mice showed hardly any change.
Fasting produced a similarly drastic response in the female mice. Normally, when food is scarce, the brain sends a message to the liver to produce glucose, conserving a minimum level in the blood. But when food was withheld from female mice missing the CRFR1 receptor, the amount of glucose their livers produced dropped significantly. In hungry male CRFR1-deficient mice, the result was similar to the effects of exposure to cold: the exchange of materials in their bodies was barely affected.
“We discovered that the receptor has an inhibitory effect on the cells, and this is what activates the sympathetic nervous system,” says Dr. Kuperman.
Among other things – revealing exactly how this receptor works and how it contributes to the stress response – the findings show that male and female bodies may exhibit significant differences in the ways that materials are exchanged under stress. Indeed, the fact that the receptor suppresses hunger in females may help explain why women are much more prone to eating disorders than men.
Because drugs can enter the hypothalamus with relative ease, the findings could be relevant to the development of treatments for regulating hunger or stress responses, including anxiety disorders or depression. Indeed, several pharmaceutical companies have already begun developing psychiatric drugs to block the CRFR1 receptor. The scientists caution, however, that because the cells are involved in the exchange of materials, blocking the receptor could turn out to have such side effects as weight gain.
Prof. Alon Chen’s research is supported by the Henry Chanoch Krenter Institute for Biomedical Imaging and Genomics; the Perlman Family Foundation, Founded by Louis L. and Anita M. Perlman; the Adelis Foundation; the Irving I Moskowitz Foundation; the European Research Council; the estate of Tony Bieber; and the Ruhman Family Laboratory for Research in the Neurobiology of Stress.The Weizmann Institute of Science in Rehovot, Israel, is one of the world’s top-ranking multidisciplinary research institutions. The Institute’s 3,800-strong scientific community engages in research addressing crucial problems in medicine and health, energy, technology, agriculture, and the environment. Outstanding young scientists from around the world pursue advanced degrees at the Weizmann Institute’s Feinberg Graduate School. The discoveries and theories of Weizmann Institute scientists have had a major impact on the wider scientific community, as well as on the quality of life of millions of people worldwide.
Newswise —
If you’re an avid runner, logging dozens of miles every week and you happen to be over 65, odds are you’re burning oxygen at nearly the same rate as a runner in her 20s.
Scientists call this rate of oxygen consumption “running economy” and a new study by HSU Kinesiology Professor Justus Ortega and his colleagues at University of Colorado, Boulder is helping define the benefits of maintaining a running habit well into one’s senior years.
The study was published recently in Medicine & Science in Sports & Exercise, the official journal of the American College of Sports Medicine.
Previous studies indicated that running economy worsens as people age because muscles imbalances develop, resulting in muscles that are working against each other across body parts, or because overall muscle efficiency decreases.
However, these studies only surveyed people up to 61 years old. This new study takes a look at runners over the age of 65 and how their bodies cope with the demands of the sport.
Researchers found that individuals who maintain an active jogging habit into their senior years are spending nearly the same amount of metabolic energy as a 20-year-old. But what about those don’t jog? What’s causing those non-active seniors to see such increases in the metabolic costs of moving?
“Our prior research suggests that the muscles themselves are becoming less efficient. I like to think of it as your body is like a car with a fuel efficiency level,” says Ortega. “Your body has its own fuel efficiency and what we’ve seen is that the fuel efficiency in muscles is reduced in older adults who are sedentary or only walk occasionally.
The present study looked at running economy and mechanics 15 young runners and 15 older runners. Each participant had a history of running at least three times per week for a minimum of 30 minutes per session over a six-month period.
The study’s trials took place on a specialized treadmill, which read the amount of force a user applies to the running deck. Participants ran 5-minute sessions, at 2.01, 2.46 and 2.91 meters per second (4.5-6.5 mph.
Researchers found that despite differences in running mechanics, older runners consumed metabolic energy at a similar rate as young runners across the range of speeds. Essentially, these older runners maintain a youthful running economy into their 60s. However, researchers did find differences in the biomechanics of the two age groups, indicating that older runners adjust their techniques as they age, but still maintain youthful energy levels while exercising.
Future studies are aimed at determining whether other choices of exercise can have the same effect on increasing muscle efficiency that running does and whether a sedentary individual can reap the same benefit if they decide to become more active.“There’s good evidence that it’s never too late to get into exercise, it’s about finding what types of exercise are right for your body,” says Ortega.
Newswise —
A large worldwide study has found that, contrary to popular thought, low-salt diets may not be beneficial and may actually increase the risk of cardiovascular disease (CVD) and death compared to average salt consumption.
In fact, the study suggests that the only people who need to worry about reducing sodium in their diet are those with hypertension (high blood pressure) and have high salt consumption.
The study, involving more than 130,000 people from 49 countries, was led by investigators of the Population Health Research Institute (PHRI) of McMaster University and Hamilton Health Sciences.
They looked specifically at whether the relationship between sodium (salt) intake and death, heart disease and stroke differs in people with high blood pressure compared to those with normal blood pressure.
The researchers showed that regardless of whether people have high blood pressure, low-sodium intake is associated with more heart attacks, strokes, and deaths compared to average intake.
“These are extremely important findings for those who are suffering from high blood pressure,” said Andrew Mente, lead author of the study, a principal investigator of PHRI and an associate professor of clinical epidemiology and biostatistics at McMaster’s Michael G. DeGroote School of Medicine.
“While our data highlights the importance of reducing high salt intake in people with hypertension, it does not support reducing salt intake to low levels.
“Our findings are important because they show that lowering sodium is best targeted at those with hypertension who also consume high sodium diets.”
Current intake of sodium in Canada is typically between 3.5 and 4 grams per day and some guidelines have recommended that the entire population lower sodium intake to below 2.3 grams per day, a level that fewer than five per cent of Canadians and people around the world consume.
Previous studies have shown that low-sodium, compared to average sodium intake, is related to increased cardiovascular risk and mortality, even though low sodium intake is associated with lower blood pressure.
This new study shows that the risks associated with low-sodium intake – less than three grams per day – are consistent regardless of a patient’s hypertension status.
Further, the findings show that while there is a limit below which sodium intake may be unsafe, the harm associated with high sodium consumption appears to be confined to only those with hypertension.
Only about 10 per cent of the population in the global study had both hypertension and high sodium consumption (greater than 6 grams per day).
Mente said that this suggests that the majority of individuals in Canada and most countries are consuming the right amount of salt.
He added that targeted salt reduction in those who are most susceptible because of hypertension and high salt consumption may be preferable to a population-wide approach to reducing sodium intake in most countries except those where the average sodium intake is very high, such as parts of central Asia or China.
He added that what is now generally recommended as a healthy daily ceiling for sodium consumption appears to be set too low, regardless of a person’s blood pressure level.
“Low sodium intake reduces blood pressure modestly, compared to average intake, but low sodium intake also has other effects, including adverse elevations of certain hormones which may outweigh any benefits. The key question is not whether blood pressure is lower with very low salt intake, instead it is whether it improves health,” Mente said
Dr. Martin O’Donnell, a co-author on the study and an associate clinical professor at McMaster University and National University of Ireland Galway, said: “This study adds to our understanding of the relationship between salt intake and health, and questions the appropriateness of current guidelines that recommend low sodium intake in the entire population.”
“An approach that recommends salt in moderation, particularly focused on those with hypertension, appears more in-line with current evidence.” The study was funded from more than 50 sources, including the PHRI, the Heart and Stroke Foundation of Canada and the Canadian Institutes of Health Research
Newswise —
During the 2014-15 flu season, the poor match between the virus used to make the world’s vaccine stocks and the circulating seasonal virus yielded a vaccine that was less than 20 percent effective.
While this year’s vaccine is a much better match to the circulating seasonal strains of influenza, the shifty nature of the virus and the need to pick the viruses used to make global vaccine stocks well before the onset of the flu season can make vaccine strain selection a shot in the dark.
That process — dependent on the careful selection of circulating virus strains and the identification of mutations in the part of the virus that recognizes host cells — could soon be augmented by a new approach. It would more precisely forecast the naturally occurring mutations that help seasonal flu virus dodge the vaccine.
Writing this week (May 23, 2016) in the journal Nature Microbiology, a team of researchers led by University of Wisconsin-Madison School of Veterinary Medicine virologist Yoshihiro Kawaoka describes a novel strategy to predict the antigenic evolution of circulating influenza viruses and give science the ability to more precisely anticipate seasonal flu strains. It would foster a closer match for the so-called “vaccine viruses” used to create the world’s vaccine supply.
The approach Kawaoka and his colleagues used involved techniques commonly employed in virology for the past 30 years and enabled his group to assemble the 2014 flu virus before the onset of the epidemic. “This is the first demonstration that one can accurately anticipate in the lab future seasonal influenza strains,” explains Kawaoka, a UW-Madison professor of pathobiological sciences who also holds a faculty appointment at the University of Tokyo. “We can identify the mutations that will occur in nature and make those viruses available at the time of vaccine (virus) candidate selection.”
Influenza depends on its ability to co-opt the cells of its host to replicate and spread. To gain access to host cells, the virus uses a surface protein known as hemagglutinin which, like a key to a lock, opens the cell to infection. Vaccines prevent infection by priming the immune system to create antibodies that effectively block the lock, prompting the virus to reengineer the hemagglutinin key through chance mutation.
“Influenza viruses randomly mutate,” notes Kawaoka. “The only way the virus can continue to circulate in humans is by (accumulating) mutations in the hemagglutinin.”
To get ahead of the constant pace of mutations in circulating flu viruses, Kawaoka’s group assembled libraries of human H1N1 and H3N2 viruses from clinical isolates that possessed various natural, random mutations in the hemagglutinin protein. The viruses were then mixed with antibodies to weed out only those that had accumulated enough mutations to evade the antibody. Because the sources of the viruses were known, the patterns of mutation could be mapped using “antigenic cartography.”
The mapping, says Kawaoka, identifies clusters of viruses featuring novel mutations which, according to the new study, can effectively predict the molecular characteristics of the next seasonal influenza virus. Such a prediction, says Kawaoka, could then be used to more effectively develop the vaccine virus stockpiles the world needs each flu season.
Each year the World Health Organization (WHO), comparing genetic sequence and antigenic data, makes recommendations about which circulating strains of influenza will make the best matching vaccine. The method described by Kawaoka and his colleagues is conceptually different in that it mimics the mutations that occur in nature and accelerates their accumulation in the critical hemagglutinin protein.
“Our method may therefore improve the current WHO influenza vaccine selection process,” Kawaoka and his group conclude in the Nature Microbiology report. “These in vitro selection studies are highly predictive of the antigenic evolution of H1N1 and H3N2 viruses in human populations.”